In this lab, you will experimentally determine how performance-critical the Linux kernel's use of prepaging is for files that are mapped into virtual memory. That is, how much worse would performance be if Linux used only the simpler approach of demand paging?
My instructions assume you will be doing this on the netbook you have been issued, running the Ubuntu Netbook Remix distribution of Linux. You will need to follow the previously-issued instructions for building a modified kernel.
The following quote from page 203 of our textbook provides some context:
... Linux ordinarily uses a variant of prepaging ... for files mapped into virtual memory. This makes sense because
reading from disk is slow. However, if the application programmer notifies the operating system that a particular memory-mapped file is going to be accessed in a "random" fashion, then Linux uses demand paging for that file's pages. The programmer
can provide this information using the madvise procedure.
This provides a clue as to how you could modify the kernel to incorporate a selectable mode in which all prepaging is disabled, resulting in pure demand paging. Namely, we just need to find out where in the kernel the "random" option from madvise exerts its influence. Somewhere there must be an if statement that in effect says "if this virtual memory area is marked as randomly-accessed, use demand paging, otherwise prepaging." You just need that to say "if this virtual memory area is marked as randomly-accessed or prepaging is disabled, use demand paging, otherwise prepaging." One of my goals on the lab preview class day is to demonstrate how you would navigate through the kernel source code, starting from the madvise system call, to find the relevant if statement to modify.
Here is an overview of what you will need to do:
Modify the aforementioned if statement to optionally disable prepaging, depending on whether a global variable you add has a zero or nonzero value.
Also modify the kernel so that the global variable is accessible through a special file in the /proc/sys/vm/ directory, so that you can control it during your experimentation.
Build and install the modified kernel.
Choose a workload for your experimentation. That is, you will need to focus on some specific use for Linux (such as running a particular program) to see how much its performance depends on prepaging.
Use a script to time lots of runs of your workload, in a randomized mix of the prepaging-disabled and prepaging-enabled conditions, flushing the page cache before each run.
Statistically summarize and assess your data, using graphical and numerical techniques to test whether prepaging made a difference, and if so, how large.
Write a clear, concise scientific report that explains what problem you addressed, the way in which you addressed it, and the results you obtained.
Once you have located the appropriate if statement, you should make two changes in that kernel source file:
Before the procedure in which the if appears, add a declaration of a new global variable, of type int which will be used as a boolean flag to determine whether prepaging should be disabled. For example, your declaration might be
int disable_prepaging;
Change the if statement's condition so that it takes the variable into account.
Now comes the question of how you will make this variable easily readable and writable. You can use a general mechanism (which I once before pointed out) that allows a kernel variable to show up as a special file. That is, you can arrange that there is a special file, let's say /proc/sys/vm/disable_prepaging, that corresponds to the global variable. If anyone looks at the current contents of that file, they will see that it contains digits corresponding to the variable's current value. (For example, if the variable is set to 0, reading from the file will return the digit 0.) Conversely, if anyone writes a numeric value into the file in the form of decimal digits, that will set the variable. A script might use this approach to set the variable with the command:
echo 1 >/proc/sys/vm/disable_prepaging
In order to make the variable show up as a file in this way, you
need to edit another kernel source file, kernel/sysctl.c.
Near the top of this file, there is a comment:
/* External variables not in a header file. */
After that comment, you can add a declaration of your variable such as
extern int disable_prepaging;
Next, look for the section of this source file where the array vm_table is initialized, which starts with the line
static struct ctl_table vm_table[] = {
You should add one more element to this array initializer; putting it at the beginning would be fine. The new element couples together the name of the file, the address of the variable, the size of the variable, and some other related information. If you have used disable_prepaging as the name for both the file and the variable, then this name will show up three times, once for the file and twice for the variable:
{
.ctl_name = CTL_UNNUMBERED,
.procname = "disable_prepaging",
.data = &disable_prepaging,
.maxlen = sizeof(disable_prepaging),
.mode = 0644,
.proc_handler = &proc_dointvec,
},
Whenever testing the performance implications of an operating system change, you need to select a workload, because the performance change might be different for different workloads. Ideally your test workload should be a realistic reflection of the kinds of work people really use the system for, but be a simple, repeatable test that doesn't take unreasonably long to execute many times. These desiderata are inherently in conflict, so you will inevitably need to choose a tradeoff. For this lab, you might consider using one of the following, or perhaps several of them run in succession:
using the g++ compiler to compile and link a C++ program, the way you did in the prior lab
running a Python script, or even just a simple one-line Python computation such as
python -c 'print 2+2'
starting up the emacs text editor and having it then immediately exit:
emacs -kill
starting up the OpenOffice.org writer and having it then immediately exit:
oowriter -terminate_after_init
You'll need to run your workload lots of times, which would be a pain to do by hand. Additionally, if you were to do it by hand, you'd need to be very careful to get all the details right of running the workload the same way each time, except for the one intentional change of turning prepaging on or off. This is made more complicated by the fact there there would be some additional commands to run beyond the workload itself:
Before each run of the workload, you'll want to put the memory back into a clean starting state. The ideal would be a reboot, but that's a time-consuming nuisance. A reasonable compromise would be to use the following two commands:
sync echo 3 >/proc/sys/vm/drop_caches
The first of these writes any dirty pages (or other file-related information, such as directory entries) out to the disk. The second eliminates from memory any cached copies of previously-accessed pages (and other file-related information).
Before each run of the workload, you'll need to set prepaging to its disabled or enabled state, using a command such as shown earlier.
Assuming that you are writing the workload execution time out to a data file, you will probably also want to write out some identifying information so that it is clear which condition each time originated from.
An additional complication is that the runs with and without prepaging should be shuffled together in a random order. That way, if there is any extraneous factor that is making some runs take longer than others, it will be equally likely to influence runs of each of the two kinds.
All of this suggests that you could benefit from a little bit of machinery to help automate your experimentation. I suggest using two levels of scripting. The actual experiment can be run using a simple shell script (to be run with the bash command) that just lists one by one all the commands to execute. However, rather than your using a text editor to type in that shell script, you can use a higher-level program to generate the shell script. This higher-level program would take care of randomization and making all the right numbers of copies of the right commands. I suggest using a Python script for this higher-level program.
Let's work an example. In the previous lab, you learned that the hard disk buffering can be turned on or off using the hdparm command. For the writer program, this had enormous performance consequences. Let's see what the performance consequences are for a somewhat more realistic workload. In particular, let's see what the impact is on unpacking the Linux source tree from its tar file and then removing the resulting directory tree. We'll use this as an example performance study not only in this section on scripting, but also in the later section on statistical analysis.
I've attached the Python script, named make-script.py, that I wrote to generate the shell script for this experiment. You would run it using a command like
python make-script.py >script.sh
This creates the shell script, script.sh. If you try this out, you may wind up with a somewhat different version that I've attached here, because the runs may be in a different random permutation. This script contains the detailed step-by-step instructions for conducting the experiment. To run it, putting the resulting timing data in a file, you would use a command like
sudo bash script.sh >buffering.csv
This grinds away a long time doing the various experimental runs and in the process creates a file, buffering.csv, that shows the resulting times in a comma-separated-values form. In this CSV file, the first column is a 0 or 1 showing whether buffering was off or on for the particular run, and the second column is the resulting execution time of the workload, in seconds. The file also starts with a header row identifying each of the columns.
You'll need to do something analogous, but instead of turning disk buffering on and off, you'll turn prepaging on and off. And you'll use the workload you chose, rather than the unpack-and-delete workload used here. You should choose for yourself how many repetitions to do under each condition. That depends on such factors as how time consuming your workload is, how consistent the times are under each of the two conditions, and how big the performance effect is that you are measuring. You may want to do some preliminary, informal, experimentation to get a feel for those factors, before you settle on the number of repetitions to use in your actual experiment.
Continuing with our example from the previous section, we have a CSV file that shows the impact of disk buffering, but a long list of numbers isn't really a very helpful form in which to present the data. We can use the R system, a free statistics package, to summarize the data in both graphical and numerical form. The R system is installed on the MCS lab macs and you can also download your own copy if you would prefer. (It is available for a variety of platforms; I was able to install it on my netbook, for example.)
Once I launched R, I entered the following two commands to read the CSV file into an R variable named bufferingData and then graphically summarize that data in a boxplot while simultaneously putting a numerical summary in the variable b:
bufferingData <- read.csv("buffering.csv")
b <- boxplot(time ~ buffering, data=bufferingData, range=0, names=c("off", "on"), main="linux source tree untar and remove", ylab="time (seconds)", xlab="hard disk buffering")
The resulting boxplot popped up in a separate window:
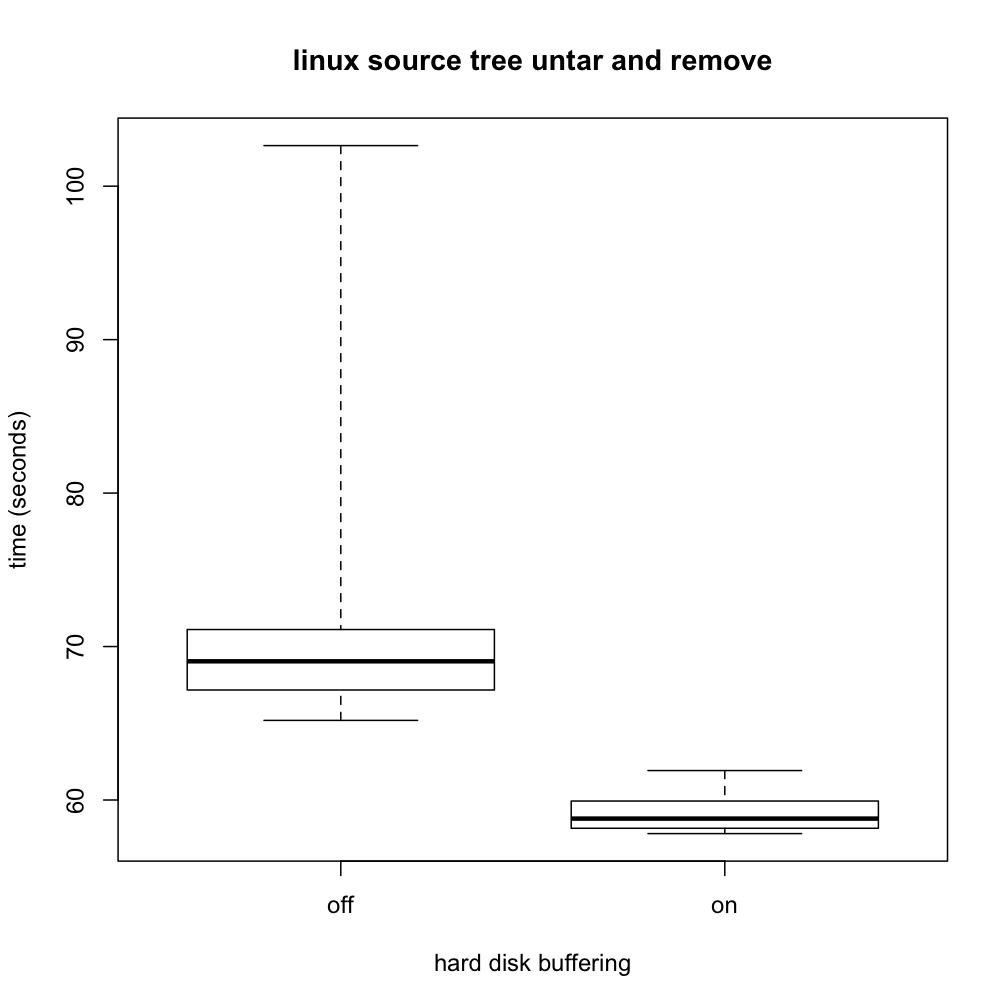
The dark horizontal line in each box indicates the median of the respective data set. You can see that the median time for the workload was larger when buffering was turned off, as you might expect, though the difference was nowhere near as large as with writer. The lighter box surrounding each median indicates the middle half of the data, that is the range from the 25th to 75th percentile. The protrusions beyond the boxes (known as "whiskers") indicate the full range, all the way from the fastest run up to the slowest. From these, you can see at a glance that all 20 runs with buffering on were faster than all 20 runs with buffering off; the ranges don't overlap at all. Even without a formal statistical test of significance, that is very strong evidence that disk buffering does affect this workload's performance, even if not so dramatically as with writer. Other facts that are apparent include that the distributions are rather asymmetrical and that they don't have the same dispersion as each other. These suggest we will need to exercise care in selecting a formal statistical test; a test that presumes equal-variance normal distributions would be badly off the mark.
This boxplot would be well worth including in a scientific report of the experiment. However, it would also be useful to provide a numerical summary of the same information. This summary was saved in the R variable named b. Simply entering b on a line by itself prints out its value, which starts with the following:
$stats
[,1] [,2]
[1,] 65.1880 57.8110
[2,] 67.1690 58.1580
[3,] 69.0390 58.7880
[4,] 71.1095 59.9335
[5,] 102.6440 61.9190
Translating this into English, we'd say that turning disk buffering off increases the median execution time from 58.7880 seconds (range: 57.8110-61.9190) to 69.0390 seconds (range: 65.1880-102.6440). The corresponding interquartile ranges are 58.1580-59.9335 and 67.1690-71.1095.
These graphical and numerical descriptive statistics go a long way towards making the experimental results understandable. However, we can also go a bit further and use statistical hypothesis testing and a confidence interval estimator to answer two questions: might the apparent effect plausibly be coincidental, and how well, with our limited data, have we pinned down the size of the effect?
Given that the data clearly isn't normally distributed, the safest approach would be to use an exact test based on ranks. For example, in our data, we saw that ranks 1-20 are occupied by times from the buffering-on condition and ranks 21-40 are occupied by times from the buffering-off condition. That would be extremely unlikely to happen by chance if the two conditions were just meaningless labels applied to data all drawn from a single distribution. In fact, the chance of it happening is exactly 1 in C(40, 20), the number of ways to choose 20 items out of 40. An equally extreme result in the other direction would be just as unlikely. Therefore, seeing such an extreme result, we can reject the idea that buffering doesn't matter, with a p value of 2/C(40,20). You can calculate this and discover just how small it is, indicating that the "null hypothesis," that there was no effect, is very implausible. That gives us reason to believe the alternative hypothesis, that disk buffering has an impact on execution time. Calculating this one p-value by hand would be easy enough, but in general, if the ranks aren't so cleanly divided, the calculation is best left to the R system. It goes by the name of the Wilcoxon test.
To load in a library of exact rank tests, you can give this command to R:
library("exactRankTests")
The first time you do this, you may get an error message that the library isn't installed on your system. In this case, you can first install it using
install.packages("exactRankTests")
and then go back to the library command. In any case, once it is loaded in, the exact Wilcoxon test can be applied to the data we loaded from the CSV file:
wilcox.exact(time ~ buffering, data=bufferingData, conf.int=TRUE)
The output is as follows:
Exact Wilcoxon rank sum test
data: time by buffering
W = 400, p-value = 1.451e-11
alternative hypothesis: true mu is not equal to 0
95 percent confidence interval:
8.443 11.630
sample estimates:
difference in location
9.8345
The p-value is listed as 1.451e-11, which is to say, 1.451×10-11, a very small number indeed (and, reassuringly, the same as 2/C(40,20)). As such, the test supports the alternative hypothesis that mu (the increase in time caused by turning buffering off) is not equal to 0. Moreover, because we provided the option conf.int=TRUE, we also get information on how well pinned down this increase is. (If we had taken more data than 20 repetitions of each condition, we might have pinned it down more closely.) With 95% confidence we can say that the increase is somewhere between 8.443 and 11.630 seconds. Our best estimate of the increase is somewhere in between, 9.8345 seconds, a value similar to the difference between the medians.
Instructor: Max Hailperin